 |  |
| PyMOL built-in ribbons | "molauto -nice ... | molscript -r > ..." |
Cartoon ribbons in PyMOL rival those of the popular Molscript/Raster3D packages, but PyMOL makes creating high quality images much easier. While PyMOL can read Molscript output directly (see the chapter on Molscript), this is no longer necessary or as convenient as utilizing PyMOL's built-in cartoon ribbon capability:
 |  |
| PyMOL built-in ribbons | "molauto -nice ... | molscript -r > ..." |
Molscript's cartoons are slightly more ideal, but PyMOL comes pretty darn close!
Note that all of the images in this section were colored using the rainbow feature (Color pop-up menu) and ray-traced with antialising enabled.
One of the advantages of PyMOL's cartoon ribbon facility is that it is easy to switch between "smoothed" versions of protein secondary structure, and "correct" renditions which portray actual main chain coordinates. Although cartoons are often used solely to represent protein structures in a schematic sense, sometimes it is desirable to combine a schematic overall picture with atomic resolution in one particular location. However, unless the cartoon track properly with alpha-carbon positions, the resulting figures will look a little silly:

In the above image, the side chains are floating off into space. Disabling "flat sheets" from the Cartoons Menu or issuing the command
set cartoon_flat_sheets, 0
will make the beta strands follow the true path of the backbone through space and give a more accurate rendition of the structure.
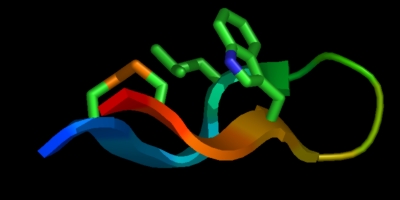
The appearance of a cartoon over the entire molecule will be substantially different when all smoothing features are turned off. For instance, with smoothing enabled:
set cartoon_flat_sheets, 1 set cartoon_smooth_loops, 1

the image differs substantially from:
set cartoon_flat_sheets, 0 set cartoon_smooth_loops, 0
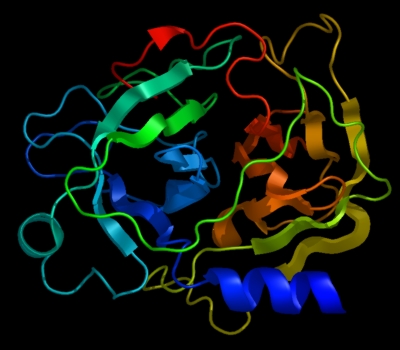
which more accurately reflects the true path of the peptide backbone:
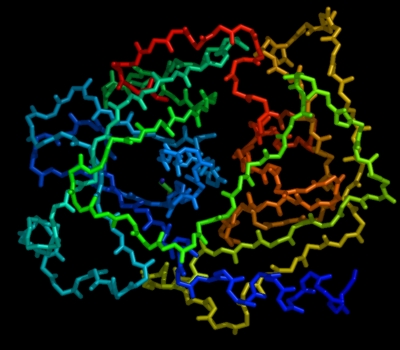
To facilitate beautiful imagery, smoothing is enabled by default (just like Molscript) [NOTE: THIS MAY CHANGE BEFORE VERSION 1.0] . Just be sure to turn it off when you want to study structures at atomic resolution (remember, real life is a bit more complicated than what you see in cartoons!).
Best results will be obtained when secondary structure information has been defined for each residue in the molecule. Under these conditions, PyMOL will do extra processing to insure that good normals have been calculated for helical regions, and perform smoothing of loops, where desired.
Also under such conditions, in automatic mode, cartoon representations will be assigned according to the secondary structure type. However, you can instruct PyMOL to ignore such information, and manually control when and where various cartoon representations are employed.
show cartoon cartoon automatic # default
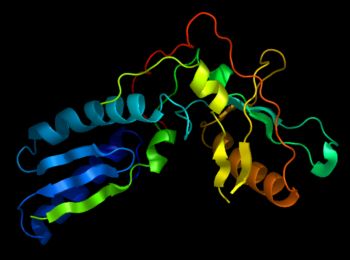
cartoon loop
cartoon rect
cartoon oval

cartoon tube
cartoon tube, 1:49/ cartoon arrow, 50:99/ cartoon loop, 100:149/ cartoon oval, 150:199/ cartoon rect, 200:250/

All cartoon ribbons have associated parameters accessible from the "set" command which allow you to change their appearance. See the chapter on Settings for more information.
set cartoon_fancy_helices, 0

Molscript addicts who simply must have those ribbon helices with tubular edges will not be disappointed with "fancy helices":
set cartoon_fancy_helices, 1

It is recommended that you read in PDB files which already have correct secondary structure assignments from a program like DSSP. However, PyMOL does have a reasonably fast secondary structure alignment algorithm called "dss". Please be aware that due to the subjective nature of secondary structure assignment in borderline cases, dss results will differ somewhat from DSSP.
SYNTAX dss selection EXAMPLE dss 1dfr
If you are visualizing an animation, you may wish derive secondary structure assignment from a specific state of the animation. This can be done with:
SYNTAX dss selection, state EXAMPLE dss mov, 1To change assignments manually, the best way is to use the alter command as follows:
show cartoon alter 11-40/, ss='H' # assign residues 11-40 as helix alter 40-52/, ss='L' # assign residues 40-52 as loop alter 52-65/, ss='S' # assign residues 52-65 as sheet alter 65-72/, ss='H' # assign residues 65-72 as helix rebuild # regenerate the cartoon