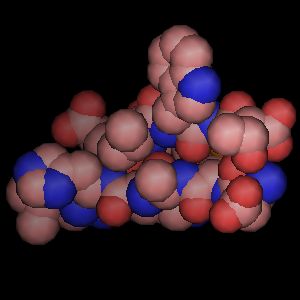
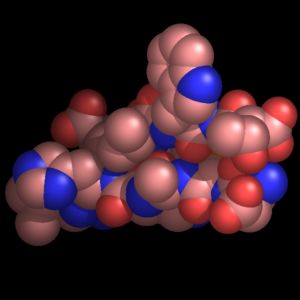
Ray-tracing produces the highest quality molecular graphics images. PyMOL is the first full-featured molecular graphics program to include a high-speed ray-tracer which works with its native internal geometries (except text).
 |  |
| OpenGL Rendering (real-time manipulation) | Ray-traced Rendering (seconds or minutes per frame) |
You can ray-trace any Scene in PyMOL by clicking the "Ray Trace" button in the external GUI or using the "ray" command. The built-in raytracer also makes it possible to easily assemble very high-quality movies in a snap.
These can be changed using the "set" command. Unless otherwise specified, the settings apply only to the ray-tracing engine and not the OpenGL renderer. Some reconciliation between the two renderers is much needed, so be warned that these settings may change in the future.
Normally, the only settings you will need to change are orthoscopic, antialias, and gamma. If you are down in an enzyme active site which is heavily shadowed, you may want to increase direct to 0.5-0.7 in order to improve brightness and contrast.
All images (ray-traced or not) can be saved in PNG format using the "png" command. This format is directly readable by PowerPoint, and can be easily converted into other formats using a package like ImageMagick. You can also save images using the "Save Image" option in the "File" menu. Images are always saved at the same resolution as the viewer window.
ray png my_image.png