In this section and those which follow, you will find complete recipes for generating figures directly from the PyMOL command language. Note that when you are using PyMOL interactively, you can manually perform the same actions using the mouse and menus instead of typing commands, and thus your scripts can be quite simple and shorter than what you see here.
WARNING: Some of these examples require significant memory and processor speed in order to be practical. I recommend a 500+ mhz machine with at least 256 MB of RAM for most of these. Less RAM is okay, but you'll end up using virtual memory (slow!).

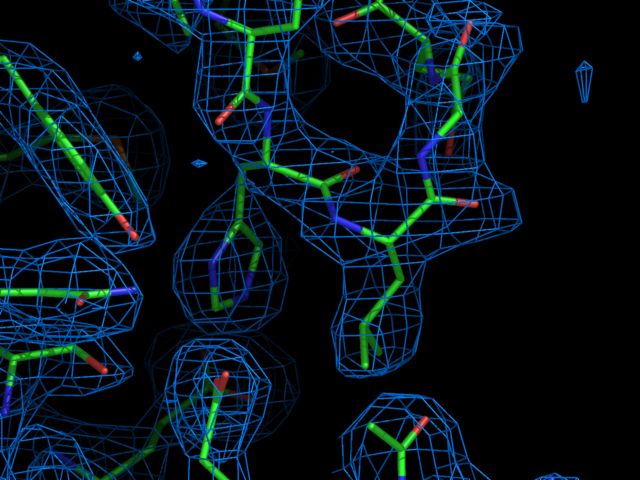
# alter settings for publication quality set mesh_radius = 0.02 set antialias = 1 set stick_radius = 0.1 # load pdb and map file load 1DN2.pdb,1dn2 load 2fofc.xplor # display region of interest hide show sticks,(byres ((c;f & i;5,6) x;4)) # show e-density nearby isomesh den, 2fofc, 1.0, (c;F & i;6), 8.0 color marine,den # zoom in, turn, and clip zoom (c;f & i;5,6),2 turn x,10 turn y,-10 clip slab, 6.5 # ray trace and write image file ray png density.png
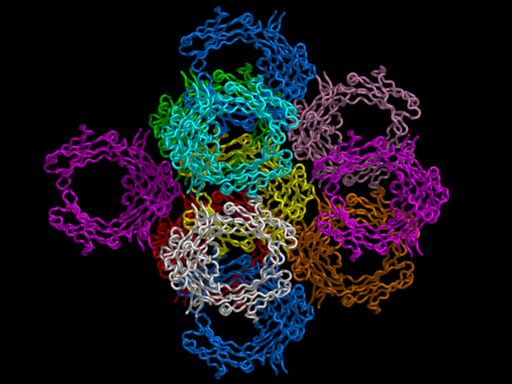
# load pdb file which has a CRYST1 record load 1DN2.pdb,1dn2 # create neighbors with contacts symexp s,1dn2,(all),4.0 # display ribbon only, and make it a little bit "fatter" hide set ribbon_radius = 0.75 show ribbon # rotate and zoom turn y,90 move z,50 zoom # color symmetry-related copies distinctly color white color yellow, (1dn2) color cyan, (s01-10100) color marine, (s0000-100 or s00000100) color red, (s01000000) color violet, (s00000001 or s000000-1) color orange, (s01000001) color pink, (s01000101) color green, (s01000100) # now render (about 1 minute) set antialias=1 ray png packing.png
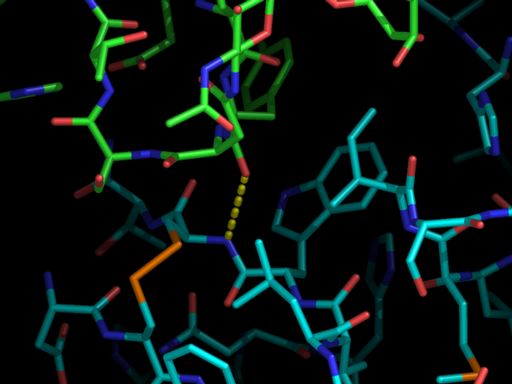
# load pdb file which has a CRYST1 record
load 1DN2.pdb,1dn2
# create neighbor with contact near a certain atom
symexp s,1dn2,(c; F and i;12 and n;N),4.0
# hide everything outside region of interest
hide (!(byres ((1dn2 and c; F and i;12 and n;N) x;12)))
# color-by-atom-cyan on 1dn2 to distinguish it from symmetry-related copy
util.cbac 1dn2
# restore view saved using from get_view in a previous session
set_view (\
-0.042664494, -0.003409438, 0.999083281,\
-0.617746830, 0.786019623, -0.023697896,\
-0.785219014, -0.618191123, -0.035642438,\
-1.056621075, -1.729980350, -39.743129730,\
29.859001160, 38.402999878, 19.087999344,\
27.443143845, 46.443149567, 1.000000000 )
# show the single hydrogen bond in the interface
dist ( s01000000 segi '' chain B resn TYR resi 296 name O ),\
( 1dn2 segi '' chain F resn CYS resi 12 name N )
# image
set antialias=1
ray
png contact.png