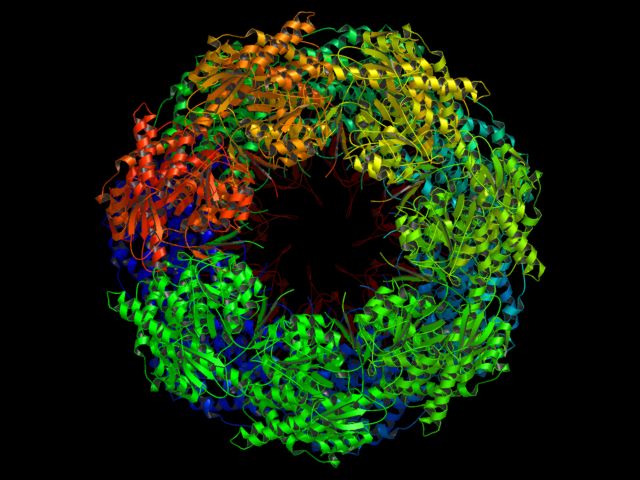
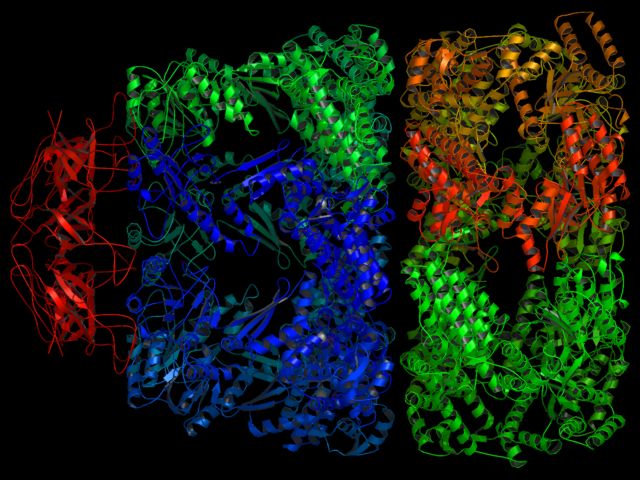
Here are some examples of things that PyMOL was never designed to do, but that it manages to accomplish anyhow!
Requires Molscript. CPU Time: about 4.5 minutes on a 1 Ghz Athlon (256 MB/Linux) for two images. Used over 300MB RAM/Swap


# run molscript on the PDB file system molauto -nocentre 1AON.pdb | molscript -r > 1AON.r3d # load raster3D input file load 1AON.r3d # zoom in a little reset move z,100 # render set antialias=1 ray png groel_es1.png # render turn y,90 ray png groel_es2.png
CPU Time: 66 seconds on a 1 Ghz Athlon (256 MB/Linux). Used over 350MB RAM/swap.
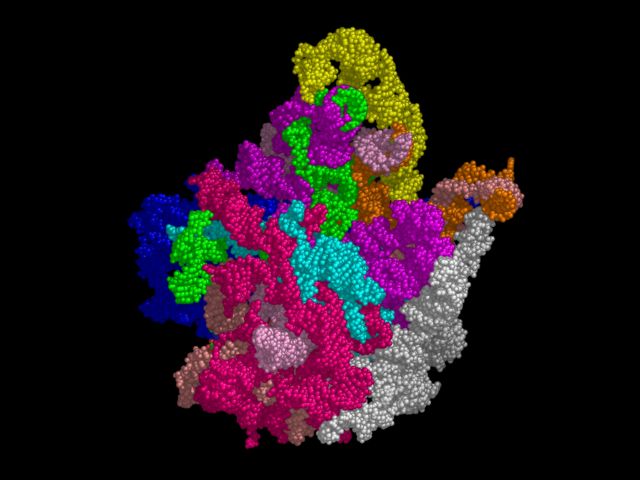
load 1FFK.pdb,1ffk # create object with only nucleic acid create nuc = (1ffk and not n;ca,cd) del 1ffk # color contiguous chains edit (c;0 & n;P & i;713) color blue,(pkchain) edit (c;0 & n;P & i;2250) color green,(pkchain) edit (c;0 & n;P & i;1876) color magenta,(pkchain) edit (c;0 & n;P & i;2596) fcolor violet,(pkchain) edit (c;0 & n;P & i;1993) color cyan,(pkchain) edit (c;0 & n;P & i;2764) color white,(pkchain) edit (c;0 & n;P & i;1552) color salmon,(pkchain) edit (c;0 & n;P & i;857) color pink,(pkchain) edit (c;0 & n;P & i;1149) color orange,(pkchain) edit (c;9 & n;P & i;21) color yellow,(pkchain) # orient the "hand" reset turn y,-60 turn z,180 move y,-15 # show spheres show spheres # ray trace set antialias=1 ray # write output png ribosome.png
CPU Time: 25 minutes on a 1 Ghz Athlon (256 MB/Linux). Used over 840 MB (!) of RAM/swap and involved raytracing over a half-million triangles. Can you imagine working with 500 Kd worth of protein surfaces in any other program?
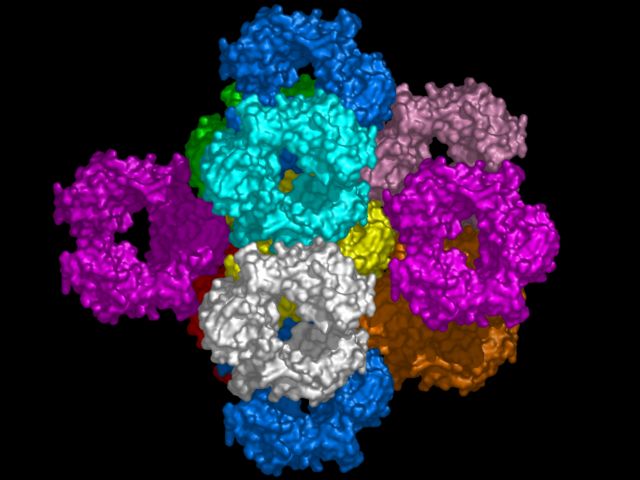
# load pdb file which has a CRYST1 record load 1DN2.pdb,1dn2 # remove waters remove (resn hoh) # create neighbors with contacts symexp s,1dn2,(all),4.0 # display surfaces hide show surface # rotate and zoom reset turn y,90 move z,30 turn x,5 turn y,5 # color symmetry-related copies distinctly color white color yellow, (1dn2) color cyan, (s01-10100) color marine, (s0000-100 or s00000100) color red, (s01000000) color violet, (s00000001 or s000000-1) color orange, (s01000001) color pink, (s01000101) color green, (s01000100) # now render set antialias=1 ray png packsurf.png